Triple-negative breast cancer, also known as TNBC, is an invasive, aggressive and hard-to-treat beast. The cancer cells lack the expression of three cell surface molecules that are common targets for therapy, and the prognosis for patients can be poor. It's left its mark on my own family, but we are far from the only ones affected.
Doctors and researchers have been struggling for years to understand more about TNBC and how best to tackle it. Although it's been known for some time that TNBC patients with higher numbers of immune cells in their tumors seem to fare better than those with fewer, it's not well understood why.
Now pathologist Michael Angelo, MD, PhD, and postdoctoral scholar Leeat Keren, PhD, have used a variation of a new imaging technique to precisely identify the type and location of immune and cancer cells with each patient's tumor.
They published their results last week in Cell.
Understanding the locations of cells within a tumor is likely to provide significant insight into how each tumor grows and responds (or doesn't respond) to therapy. As Angelo explained in an email to me:
Cells constantly communicate with each other. They integrate information from their surroundings and respond to signals from their immediate environment. Obtaining information about the environment of the cell can point towards the mechanisms driving its behavior.
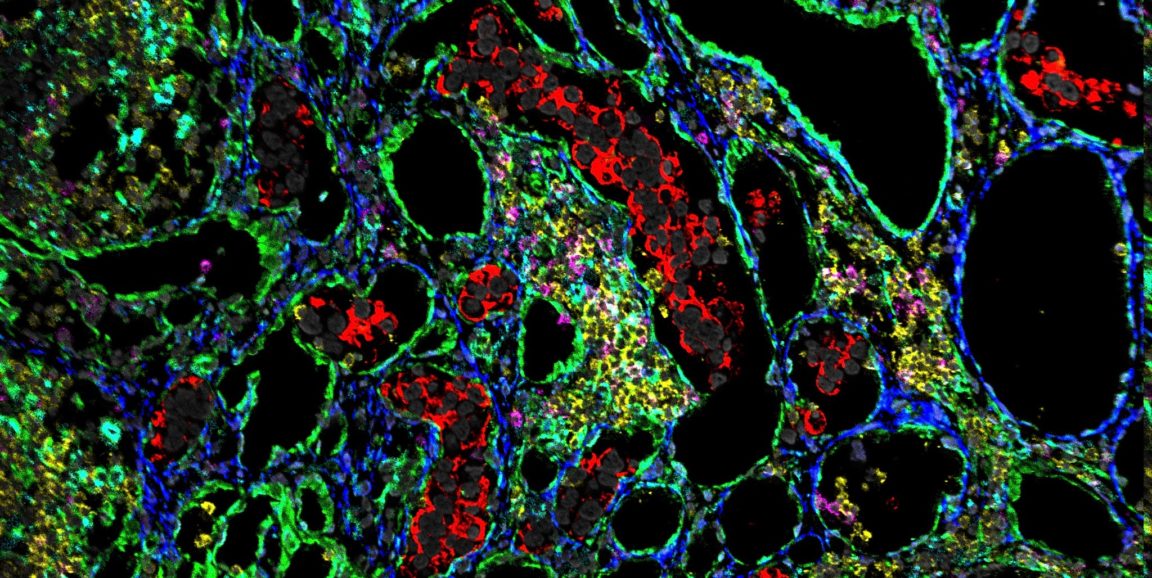
Angelo and Keren used a technique, called multiplexed ion beam imaging by time of flight, or MIBI-TOF, that can track the expression of up 40 proteins in a single tissue sample. Until recently it was only possible to see three or four proteins at a time on one slide. In contrast, as shown in the photo above, MIBI-TOF showcases the location of many proteins in a dazzlingly colorful array that reveals the type, location and even behavior of each cell in the tumor. (In the photo, the tumor cells are red and the immune cells are pink, yellow and cyan.)
They used tissue samples from 41 TNBC patients to sort patient samples into two categories: one in which the immune cells had infiltrated the tumor microenvironment, which they designated as "mixed," and the other in which the immune cells crowded along defined borders outside and within the tumor, which they called "compartmentalized." These differences correlated with patient survival, they found.
As Keren described:
It has been known for a while that the number of immune cells in the tumor is related to patient survival in TNBC. Here we found that even for patients with similar numbers of immune cells, the identity of these immune cells and their organization within the tissue varied greatly between patients and had an even greater association with prognosis. We found that patients whose tumors were compartmentalized were significantly more likely to survive their cancers than those whose tumors were mixed.
But what was really remarkable was that these groups were also associated with cell populations that expressed different immunoregulatory proteins. For example, in the mixed group, the immunosuppressive protein PD1 was primarily expressed by immune cells known as cytotoxic T cells, whereas in the compartmentalized group it was primarily expressed by helper T cells. This is an exciting new link of how microscopic expression patterns of proteins can coincide with macroscopic phenomena such as cell organization within the tumor and patient survival.
We've written here before about variations on this imaging technique, which was pioneered in the laboratory of Stanford microbiologist and immunologist Garry Nolan, PhD. It's so exciting to witness how researchers are using it to understand more about the geography of cancers. But much still remains to be learned.
The researchers are now hoping to use the technique to investigate responses to immunotherapy and to better understand how the immune composition of tumors changes over time. The opportunities abound, Angelo said:
A big advantage of MIBI-TOF is that it is can be used on clinical samples from long-term storage. This means that it can be used to interrogate around 1 billion samples stored in clinical repositories around the world. This opens up a wealth of information to researchers that until now has been largely untapped.
Photo by Leeat Keren