Computers have revolutionized many fields, so it isn't surprising that they may be transforming cancer research. Computers are now being used to model the molecular and cellular changes associated with individual tumors, allowing scientists to simulate the tumor's response to different combinations of chemotherapy drugs.
Modeling big data to improve personalized cancer treatment was the focus of a recent episode of the Sirius radio show "The Future of Everything." On hand was Sylvia Plevritis, PhD, a professor of biomedical data science and of radiology at Stanford, who discussed her work with Stanford professor and radio show host Russ Altman, MD, PhD.
Plevritis and her colleagues are using multi-omics data -- including measures of gene expression, protein function, metabolic processes and more -- to extensively profile individual tumors of individual patients.
They are analyzing this data to better understand how tumors become drug-resistant. She explained in the podcast that tumors are often heterogeneous -- not every cell has the same gene mutations -- but chemotherapy drugs typically target specific genetic mutations. Tumors are also driven by complex mechanisms beyond genetic mutations. So her lab is comprehensively characterizing the different cell types in a tumor and how these different cell types respond to individual drugs. By better understanding the complexity of what drives the tumor's response, they hope to identify the underlying mechanisms of drug resistance.
The goal, Plevritis said, is to more accurately estimate the response of the entire tumor to a given set of drugs without having to run clinical trials on every drug combination. Using their modeling, they hope to identify the most promising drug combinations to make clinical trials more efficient, she said.
The research team tested their computational model by measuring the multi-omics profile of human cancer cells in a dish, before and after exposing the cells to specific drugs. Their model then identified the minimum combination of drugs with the maximum effect. This work used archived cell samples, so their modeling results didn't impact the patients' treatment. But they compared their model's prediction to what drugs the patients actually received.
They determined that the best chemotherapy cocktail for most of the patients would have been just one or two of the drugs that they received. For about 10 percent of the patients, they predicted that a totally different drug would have been the most effective, Plevaritis said in the podcast.
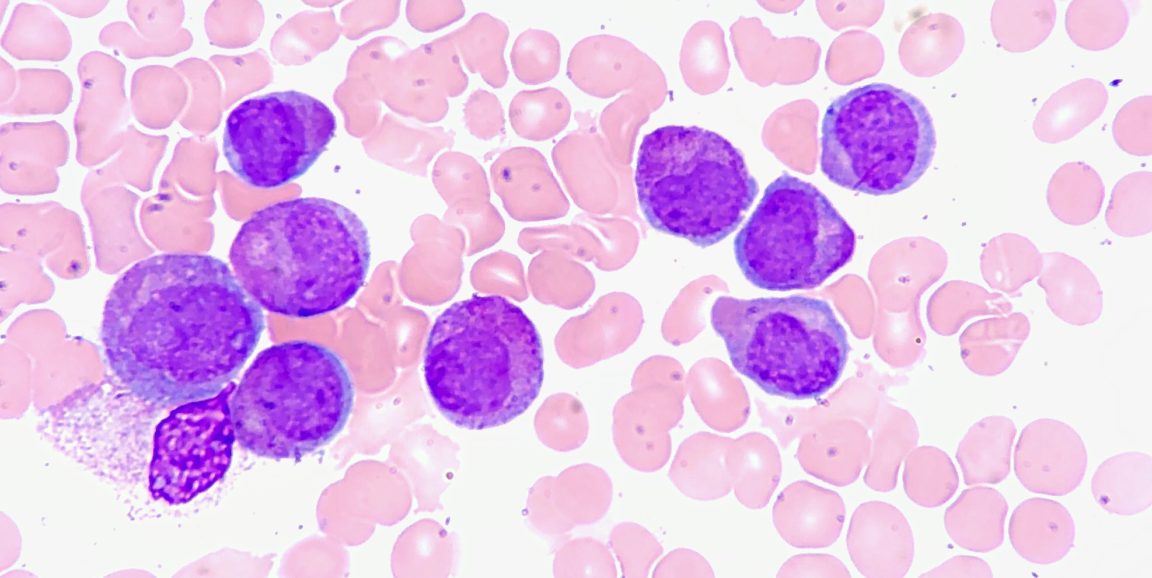
Thus, their computational model may be able to divide patients into different groups, based on tumor characteristics, and match those groups with specific chemotherapy cocktails that would be most effective for them. Plevaritis' team is currently setting up a study to validate their computational predictions for a group of patients with acute myeloid leukemia, in parallel with a combination drug therapy trial, she said.
As a member of the Cancer Intervention Surveillance Network Modeling consortium, Plevritis is also using computational models to evaluate the impact of cancer screening guidelines -- such as the recommended frequency of mammograms for general breast cancer screening -- on mortality rates. For example, policy organizations like the U.S. Preventive Services Task Force often ask the consortium to simulate thousands of different screening policies -- and rank their potential impact -- to use as part of their selection criteria, she said.
One outcome of this work is an online decision tool for women who are at high risk for developing breast cancer because they carry a mutation in the BRCA1 or BRCA2 gene. Plevritis said about 45,000 people worldwide have used the tool, and her team has received a lot of positive feedback.
"It's been very satisfying to get these emails and this feedback from individuals who feel that this complex information was distilled in a way that they can make sense of it," Plevritis said.
Image of acute promyelocytic leukemia cells by Ed Uthman