The Health After Cancer podcast brings together Stanford Medicine physicians, cancer survivors and advocates to discuss issues around cancer survivorship and health after cancer treatment.
Author: Krista Conger
Do you sound like you? Gender-affirming voice therapy allows people to speak authentically
Why is it important to offer gender-affirming voice therapy or surgery? We spoke with experts on all sides of the equation.
What’s the deali-O Part 2: Navigating new weight loss drugs
Stanford Medicine obesity experts discuss the pros and cons of semaglutide, the active ingredient in Ozempic and other weight loss drugs.
This Is My Why: LGBTQ+ education
Stanford Medicine researchers, health care workers and staff speak to why they're passionate about LGBTQ+ care, education and advocacy.
What’s the deali-O with new weight loss drugs? Part 1
Stanford Medicine researchers weigh in on the promise and peril of increasingly popular diabetes drugs being used for weight loss.
A network to support LGBTQ+ health
Researchers at Stanford Medicine are investigating how to best provide health care to the LBGTQ+ community.
Ribosomes’ sweet spot
Researchers at Stanford Medicine discover an unknown quirk about the ribosome and make a sweet representation along the way.
Molecules, shmolecules. Why should you care?
We explore the most basic molecular elements of human biology in the lead story for the latests issue of Stanford Medicine magazine.
Antibiotics might not be the best answer for C. diff
A new testing tool helps doctors avoid unnecessary antibiotic treatment in patients with diarrhea and suspected C. diff infection.
Coming full circle with extrachromosomal DNA, cancer and Ptolemy
Research into the destructive influence tiny DNA circles have on cancer presents endless ideas for clearly describing groundbreaking science.
Tackling typhoid, one finger prick at a time
Stanford infectious disease experts devise a way to use finger-prick blood samples from small groups to detect typhoid in large populations.
Researchers discuss health disparities in melanoma diagnoses
While melanoma rates have leveled off for most of the United States, Black and Latino communities are at a higher risk for the disease.
Endemic: What it might mean for masks, COVID-19 shots and more
Stanford experts discuss what ‘endemic’ means in the context of biology, masking, vaccines, and mental and behavioral health.
Vaccines provide broad protection against COVID-19, scientists say
According to a Stanford study, vaccines against COVID-19 are better than infection at generating antibodies to recognize new viral variants.
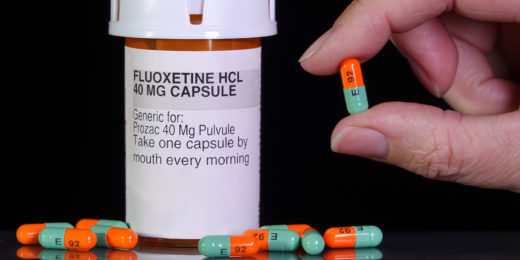
Can Prozac fight brain cancer?
The common antidepressant Prozac melts away glioblastoma tumors in laboratory mice, suggesting possible treatment for the deadly cancer.
Who’s on first? Duking out scientific paper authorship order
Determining the order of authors on a scientific paper can be tricky. Unless you're a pair of video gaming graduate students.