Research is showing that advanced methods of genetic testing aren’t equally useful for everyone: They’re less accurate for non-white families, raising concerns about how historical gaps in whose DNA gets studied produce inequities in medical care.
Author: Erin Digitale
Why precision medicine’s targeted interventions may help prevent dangerously early births
Approximately 10% of babies worldwide are born three or more weeks before their due date -- making premature birth the leading cause of death for children under 5 globally.
How California is taking on inequity for Black patients during pregnancy, childbirth
Across the United States, Black women are three to four times as likely as their white peers to experience life-threatening pregnancy complications or die giving birth
More kids are being hospitalized for eating disorders — researchers learned why
Over the last decade, physicians have taken a broader view of adolescent eating disorders, thanks to a growing recognition of the variety of disordered eating patterns that can harm patients’ health, especially their heart function.
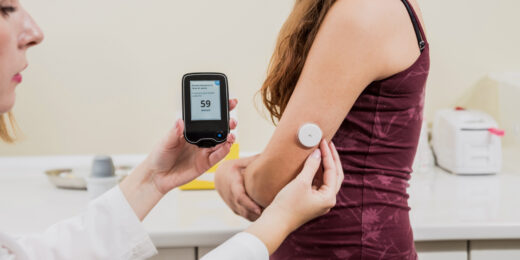
Why precision medicine leads to better diabetes care
Improvements in treatment technology are helping physicians deliver individualized care to their Type 1 diabetes patients.
No more ‘just deal with it’: Helping teens have easier periods
Nichole Tyson, MD, has advice for young people seeking help for menstrual problems – including painful, irregular or heavy periods.
Stanford Medicine making strides in advancing women leaders
Stanford Medicine is celebrating Women in Medicine month and highlighting our high rate of women in high-level roles.
How to safeguard teens’ well-being on social media
A Stanford Medicine expert has tips for parents wondering how to help teens balance mental health concerns and social media use.
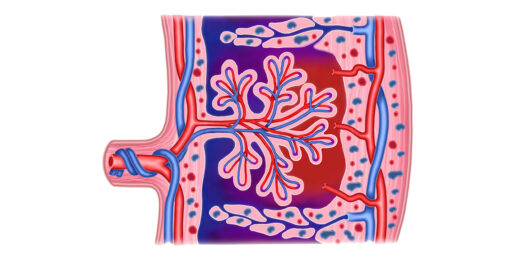
Stanford Medicine researchers map morphing placenta
Researchers at Stanford Medicine have created a detailed map of how cells in the placenta change during pregnancy.
Contributing to greener Stanford Medicine operating rooms
Stanford Medicine sustainability experts work with researchers, doctors and environmental professionals to ensure greener operating rooms.
Preparing schools for a greener future
Stanford Medicine experts, nonprofit leaders, policy makers and others "green" schools and prepare them for a climate resilient future.
Photos shine light on pediatric medical experience
Pediatric patients and their parents capture their experiences at Lucile Packard Children's Hospital Stanford, sharing photos in an exhibit.
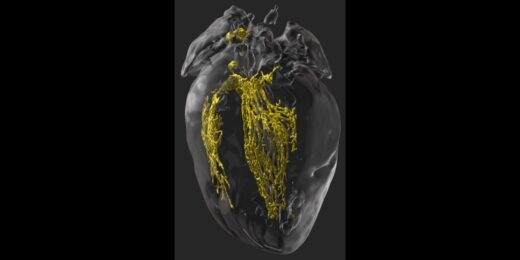
Making the invisible visible to improve heart surgery outcomes
Scientists find a way in mice to illuminate the cardiac conduction system during surgery to prevent unintended damage to healthy tissue.
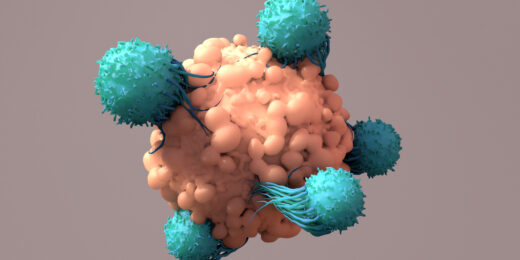
Inside-out engineering yields better cancer-fighting cells
Stanford Medicine researchers take a unique approach to refine engineered immune cells meant to kill cancer.
Kids’ storybooks encourage more water, less sugary drinks
Researchers at Stanford Medicine are finding creative ways to entice kids into drinking more water and less sugary drinks.
Revamping WHO preemie care guidelines
The new guidelines focus on parents' role in the care of vulnerable babies, as well as low-cost, evidence-based therapies.